SE(3) Score-Matching Diffusion Model
Not all data live in the Euclidean vector space. Respecting underlying symmetries of the data generally limits the model space, allowing more efficient learning and use of the data. In particular, the SE(3) group is important for modeling positions and orientations of systems ranging from protein backbones, rotamers, drones and robot arms.
This SE(3) diffusion tutorial aims to build a score-based diffusion generative models for the SE(3) roto-translational data. We will utilize trimesh and plotly packages for visualization, theseus package for SO(3) group operations because of its seamless support on pytorch’s auto-differentiation.
Table of Content
- Lie group, Lie algebra and Vector Space representations
- Recap on score-based diffusion model with an image example
- Components for diffusion in SE(3)
- Train diffusion models for 3 examples
0. Libraries and helper functions
Libraries:
apt-get -qq install libsuitesparse-dev
pip install -qq trimesh plotly theseus-aiHelpful functions:
# import the required packages
import torch, trimesh
import numpy as np
import matplotlib.pyplot as plt
from torch import nn
from tqdm.auto import tqdm
from trimesh import viewer
from IPython import display
from torch.autograd import grad
from theseus.geometry.so3 import SO3
import plotly.graph_objects as go
from plotly.express.colors import sample_colorscale
# plot the SO(3) distributions
X0 = torch.tensor([0., 0., 1.]) # using the z axis to show the rotations
def plot_rotations(rotmats, x0=X0, labels=None, cmap='jet'):
'''Plot the rotation matrices contained in a list: rotmats
colored coded by the order in the list
'''
x = np.linspace(0, 1, len(rotmats))
c = sample_colorscale(cmap, list(x))
d = []
for i, rotmat in enumerate(rotmats):
rotated_x0 = rotmat @ x0
d.append(go.Scatter3d(name=labels[i] if labels is not None else f'{i}',
x=rotated_x0[:, 0],
y=rotated_x0[:, 1],
z=rotated_x0[:, 2],
marker=dict(size=2, opacity=0.75, color=c[i]), mode='markers', ))
d.append(go.Scatter3d(name='x0',
x=x0[0:1],
y=x0[1:2],
z=x0[2:],
marker=dict(size=5, opacity=1.0, color='black'), mode='markers'))
fig = go.Figure(data=d)
fig.update_layout(width=800, height=800)
fig.show()
return None
# visualize the angle and positions of SE(3) as robot grasps or forks
def create_gripper_marker(color=[0, 0, 255], tube_radius=0.001, sections=6, scale=1.):
"""Create a 3D mesh visualizing a parallel yaw gripper. It consists of four cylinders.
Args:
color (list, optional): RGB values of marker. Defaults to [0, 0, 255].
tube_radius (float, optional): Radius of cylinders. Defaults to 0.001.
sections (int, optional): Number of sections of each cylinder. Defaults to 6.
Returns:
trimesh.Trimesh: A mesh that represents a simple parallel yaw gripper.
"""
cfl = trimesh.creation.cylinder(
radius=0.002*scale,
sections=sections,
segment=[
[4.10000000e-02*scale, -7.27595772e-12*scale, 6.59999996e-02*scale],
[4.10000000e-02*scale, -7.27595772e-12*scale, 1.12169998e-01*scale],
],
)
cfr = trimesh.creation.cylinder(
radius=0.002*scale,
sections=sections,
segment=[
[-4.100000e-02*scale, -7.27595772e-12*scale, 6.59999996e-02*scale],
[-4.100000e-02*scale, -7.27595772e-12*scale, 1.12169998e-01*scale],
],
)
cb1 = trimesh.creation.cylinder(
radius=0.002*scale, sections=sections, segment=[[0, 0, 0], [0, 0, 6.59999996e-02*scale]]
)
cb2 = trimesh.creation.cylinder(
radius=0.002*scale,
sections=sections,
segment=[[-4.100000e-02*scale, 0, 6.59999996e-02*scale], [4.100000e-02*scale, 0, 6.59999996e-02*scale]],
)
tmp = trimesh.util.concatenate([cb1, cb2, cfr, cfl])
tmp.visual.face_colors = color
tmp.visual.vertex_colors = color
return tmp
# Visualize a set of SE(3) elements H (..., 4, 4)
# by creating forks for each fork from the above function
def visualize_grasps(Hs, scale=1., p_cloud=None, energies=None, colors=None, mesh=None, show=True):
# Set color list
if colors is None:
if energies is None:
color = np.zeros(Hs.shape[0])
else:
min_energy = energies.min()
energies -=min_energy
color = energies/(np.max(energies)+1e-6)
# Grips
grips = []
for k in range(Hs.shape[0]):
H = Hs[k,...]
if colors is None:
c = color[k]
c_vis = [0, 0, int(c*254)]
else:
c_vis = list(np.array(colors[k,...]))
grips.append(
create_gripper_marker(color=c_vis, scale=scale).apply_transform(H)
)
# Visualize grips and the object
if mesh is not None:
scene = trimesh.Scene([mesh]+ grips)
elif p_cloud is not None:
p_cloud_tri = trimesh.points.PointCloud(p_cloud)
scene = trimesh.Scene([p_cloud_tri]+ grips)
else:
scene = trimesh.Scene(grips)
if show:
scene.show()
else:
return scene1. Lie Group, Lie Algebra and Vector space representations for SO(3)
In this section, we will explore the SO(3) group using the following operations. The figures are from A micro Lie theory for state estimation in robotics.

Here is a more visual or conceptual conversions.

For SO(3), group elements are 3x3 rotation matrices on the right, belonging to the manifold.
For the Lie Algebra and axis-angle representations are vectors (or can be represented as vectors) on the left, belonging to the tangent space at Identity.
-
The
expandlogare algebric operations that convert from SO(3) to so(3). -
The
veeandhatare trivial operations, extracting or constructing between axis-angle vector to skew-symmetric matrices. -
The
ExpandLogare shortcut transformations that conveniently map between SO(3) to axis-angle vectors.
torch.manual_seed(37)
ATOL, RTOL = 1e-5, 1e-5
# Dummy rotations for visualization of a sphere
R_dummy = SO3().exp_map(1.5 * torch.randn(500, 3)).to_matrix()
# v or v_vee is the axis-angle vector representation, in R^3
# v_hat is the lie algebra, in so(3) or skew symmetric matrix
# R is the 3D rotation matrix, in SO(3)
# initialize vector or axis angle representation
v1 = torch.randn(1, 3)
v2 = torch.randn(1, 3)
# rotation matrices from Exp
R1 = SO3().exp_map(v1).to_matrix()
R2 = SO3().exp_map(v2).to_matrix()
# skew symmetric matrix in so(3)
v1_hat = SO3().hat(v1)
v2_hat = SO3().hat(v2)
print('Example of the so3 skew symmetric matrix:')
print(v1_hat)
# check the consistency between vector and rotation
# 1 + 2 cos(theta) = trace(R), where theta is the length of the vector
assert torch.allclose(1. + 2. * torch.cos(v1.norm()),
torch.diagonal(R1, dim1=-1, dim2=-2).sum(), atol=ATOL)
assert torch.allclose(1. + 2. * torch.cos(v2.norm()),
torch.diagonal(R2, dim1=-1, dim2=-2).sum(), atol=ATOL)
# 1. check the vee operator
v1_vee = SO3().vee(v1_hat)
v2_vee = SO3().vee(v2_hat)
assert torch.allclose(v1, v1_vee, atol=ATOL, rtol=RTOL)
assert torch.allclose(v2, v2_vee, atol=ATOL, rtol=RTOL)
# 2. the lowercase exp is the matrix exponential from so3 to SO3
assert torch.allclose(R1, torch.matrix_exp(v1_hat), atol=ATOL, rtol=RTOL)
assert torch.allclose(R2, torch.matrix_exp(v2_hat), atol=ATOL, rtol=RTOL)
# 3. the exp_map is the uppercase Exp, from R^3 to SO3
# we used the SO3().exp_map previously but let's check
# Exp[.] = exp( hat(.) )
assert torch.allclose(R1, torch.matrix_exp(SO3().hat(v1)), atol=ATOL, rtol=RTOL)
assert torch.allclose(R2, torch.matrix_exp(SO3().hat(v2)), atol=ATOL, rtol=RTOL)
# 4. the log_map is the uppercase Log_map from SO(3) to R^3
assert torch.allclose(v1, SO3(tensor=R1).log_map(), atol=ATOL, rtol=RTOL)
assert torch.allclose(v2, SO3(tensor=R2).log_map(), atol=ATOL, rtol=RTOL)
# 5. the log operation: log R = v_hat = theta * (R - R^T) / (2 sin(theta))
assert torch.allclose(v1_hat, v1.norm() * (R1 - R1.transpose(-1, -2)) / 2. / torch.sin(v1.norm()),
atol=ATOL, rtol=RTOL)
assert torch.allclose(v2_hat, v2.norm() * (R2 - R2.transpose(-1, -2)) / 2. / torch.sin(v2.norm()),
atol=ATOL, rtol=RTOL)
# Outputs:
# Example of the so3 skew symmetric matrix:
# tensor([[[ 0.0000, -0.5821, 1.2653],
# [ 0.5821, 0.0000, -0.7207],
# [-1.2653, 0.7207, 0.0000]]])Next, we will define and check operations in the SO(3) group.
# Operation for the SO(3) group
# 1. hat(v1 + v2) = hat(v1) + hat(v2)
# because v and v_hat are in the same vector space
assert torch.allclose(v1_hat + v2_hat, SO3().hat(v1 + v2), atol=ATOL, rtol=RTOL)
# 2. compose the rotation by right multiplication
R3 = R1 @ R2
assert torch.allclose(R3, torch.einsum('...ij,...jk->...ik', R1, R2), atol=ATOL, rtol=RTOL)
# 3. compose the vector:
# The rotation composition is not commutative, i.e. R1 @ R2 != R2 @ R1
# so apparently, Exp_map(v1 + v2) != R1 @ R2.
# The reason is that the geodesic of Exp_map(v1) is different from geodesic of Exp_map(v2)
# To get the vector representation from v1 and v2, we will need to
# - compose the corresponding rotations
# - transform it back by taking the Log_map
def compose_rotvec(v1, v2):
R1 = SO3().exp_map(v1).to_matrix()
R2 = SO3().exp_map(v2).to_matrix()
R3 = R1 @ R2
return SO3(tensor=R3).log_map()
v3_a = compose_rotvec(v1, v2)
v3_b = compose_rotvec(v2, v1)
assert not torch.allclose(v3_a, v3_b)
assert torch.allclose(R1 @ R2, SO3().exp_map(v3_a).to_matrix(), atol=ATOL, rtol=RTOL)
assert torch.allclose(R2 @ R1, SO3().exp_map(v3_b).to_matrix(), atol=ATOL, rtol=RTOL)
# 4. Interpolations between two rotations on the SO3 geodesic
# See: https://en.wikipedia.org/wiki/Slerp
def slerp(R1, R2, weights):
assert R1.shape[0] == R2.shape[0] == weights.shape[0]
assert (weights <= 1.0).any() and (weights >= 0.0).any()
R1_norm = R1 / torch.norm(R1, dim=1, keepdim=True)
R2_norm = R2 / torch.norm(R2, dim=1, keepdim=True)
omega = torch.acos((R1_norm * R2_norm).sum(1))
so = torch.sin(omega)
res = (torch.sin((1.0 - weights[..., None]) * omega) / so).unsqueeze(1) * R1 + (torch.sin(weights[..., None] * omega) / so).unsqueeze(1) * R2
return res
assert torch.allclose(slerp(R1, R2, torch.tensor([1.0])), R2, atol=ATOL, rtol=RTOL)
assert torch.allclose(slerp(R1, R2, torch.tensor([0.0])), R1, atol=ATOL, rtol=RTOL)
torch.manual_seed(42)
w = torch.rand(1)
assert torch.allclose(slerp(R1, R2, w), slerp(R2, R1, 1. - w), atol=ATOL, rtol=RTOL)
# 5. "Scale" a rotation by geodesic interpolation
# Scaling operator can be done with a weighted interpolation between identity and the rotation
# 0 <= scale <= 1
def scale_rotations(R, scale):
assert len(R.shape) == 3
assert R.shape[0] == scale.shape[0]
assert (scale <= 1.0).any() and (scale >= 0.0).any()
batch = R.shape[0]
return slerp(torch.eye(3)[None, ...].repeat(batch, 1, 1), R, scale) # just use SLERP
assert torch.allclose(R1, scale_rotations(R1, torch.ones(1)), atol=ATOL, rtol=RTOL)
assert torch.allclose(torch.eye(3)[None, ...], scale_rotations(R1, torch.zeros(1)), atol=ATOL, rtol=RTOL)
# 5. "Scale" a rotation by scaling the angle while fixing the axis
# Scaling a rotation by 0 = Identity (zero degree)
# Scaling a rotation by 0.5 = rotate (0.5 * theta) wrt the same axis
# Scaling a rotation by 1.0 = the same ratation
# Therefore, again, 0 <= scale <= 1
def scale_rotations_by_angle(R, scale):
assert len(R.shape) == 3
assert R.shape[0] == scale.shape[0]
assert (scale <= 1.0).any() and (scale >= 0.0).any()
v = SO3(tensor=R).log_map() # convert from R to v
scaled_v = v * scale[..., None] # scale v, changing the theta while keeping the axis the same
return SO3().exp_map(scaled_v).to_matrix() # convert it back to the rotation matrix by Exp
# 6. Visualizing scaled rotations and interpolated rotations
n_scale = 1000
scales = torch.linspace(0.0, 1.0, n_scale + 1)
# scaling
rots = [scale_rotations(R1, torch.tensor([scale])) for scale in scales]
plot_rotations([R_dummy] + rots)
# interpolating R1 -> R2
rots = [slerp(R1, R2, torch.tensor([scale])) for scale in scales]
plot_rotations([R_dummy] + rots)
# interpolating R2 -> R1
rots = [slerp(R2, R1, torch.tensor([scale])) for scale in scales]
plot_rotations([R_dummy] + rots)The scaling is from 0.0 to 1.0, corresponding to an interpolation on the sphere from I to R (blue to red).
The interpolations between R1 and R2 are as follows:
The trajectories are the same but flipped in direction.
2. Recap on Score-based generative models
Given a ground truth \(x(0)\) and some data at time t \(x(t) \sim p_t \sim N(m(t)x(0), v(t)I)\), we can compute the score, or
\[\nabla_{x(t)}\log p_t = -\frac{x(t)-m(t)x(0)}{v(t)}\]In many of the score-matching models, the problem was designing \(m(t)\) and \(v(t)\). Here just for the demo of the score concept, we chose \(m(t) = 1\) and linear scheduling for the variance \(v(t)\).
We will use the Langevin dynamics for generation or sampling:
\[x(t+dt) = x(t) + [\nabla_{x(t)}\log p_t]dt + \sqrt{2dt}z\]where \(z\sim N(0, I)\).
- The annealed Langevin dynamics corresponds to varying \(v(t)\)
- The ODE-like sampling is the process where \(z=0\). Note that this is not the ODE-flow that aims to approximate the NLL of the data.
Some researches used SDE to design the mean and variance because there are exact form other than Langevin dynamics when we are doing the generation, which some nice properties might arise. This is out of the scope of this notebook.
import requests
from PIL import Image
from torchvision.transforms import Compose, ToTensor, Lambda, ToPILImage, CenterCrop, Resize
url = 'http://images.cocodataset.org/val2017/000000039769.jpg'
image = Image.open(requests.get(url, stream=True).raw)
print('Original Image size = ', ToTensor()(image).shape)
image_size = 128
# a series of transformations
transform = Compose([Resize(image_size),
CenterCrop(image_size),
ToTensor(), # turn into torch tensor of shape (Height x Width x Channel), divide by 255 in RGB
Lambda(lambda t: (t * 2) - 1)])
# inverse transform
reverse_transform = Compose([Lambda(lambda t: (t + 1) / 2),
Lambda(lambda t: t.permute(1, 2, 0)), # C x H x W to H x W x C
Lambda(lambda t: t * 255.0),
Lambda(lambda t: t.numpy().astype(np.uint8)),
ToPILImage()])
x_start = transform(image)
print('New Image Size = ', x_start.shape)
x_start = x_start[None, ...]
print('Batched New Image Size = ', x_start.shape)
reverse_transform(x_start[0]) # 2 sleeping cats : )
# Outputs:
# Original Image size = torch.Size([3, 480, 640])
# New Image Size = torch.Size([3, 128, 128])
# Batched New Image Size = torch.Size([1, 3, 128, 128])
# compute the scores
# here because we have access to the single ground truth, we can calculate it directly
# But in the ML problem, we want a NN to learn this
# Note that the function or model inputs are noised_x and time (or std) (we won't have ground truth during sampling)
# and output is an image of vector fields or the score
def compute_score_image(x_noised, x_truth, std, eps=1e-6):
assert x_noised.shape == x_truth.shape
assert x_noised.shape[0] == std.shape[0]
dist = (x_noised - x_truth) # (b, c, w, h)
return - dist / (std[..., None, None, None] ** 2 + eps)
# following the score back to the sample using Langevin dynamics
# noise_on = False: ODE-like or noise-free
# std_min = std_max: unannealed
def follow_score_image(x_truth, std_min, std_max, T=1500, dt=0.001, noise_on=False, seed=1001):
assert std_min <= std_max
std = torch.linspace(std_max, std_min, T + 1)
torch.manual_seed(seed)
x_noised = torch.randn_like(x_truth)
x = x_noised.clone()
for i in range(T + 1):
score = compute_score_image(x, x_truth, torch.tensor([std[i]])) # this will be where the NN model comes in
x += dt * score
if noise_on and i <= T:
x += np.sqrt(2 * dt) * torch.randn_like(x)
return x
# some visualizations
print('Un-annealed ODE-like')
x = follow_score_image(x_start, 0.5, 0.5, noise_on=False)
display.display(reverse_transform(x[0]))
print('Annealed ODE-like')
x = follow_score_image(x_start, 0.1, 5.0, noise_on=False)
display.display(reverse_transform(x[0]))
print('Un-annealed Langevin dynamics')
x = follow_score_image(x_start, 0.5, 0.5, noise_on=True)
display.display(reverse_transform(x[0]))
print('Annealed Langevin dynamics')
x = follow_score_image(x_start, 0.1, 5.0, noise_on=True)
display.display(reverse_transform(x[0]))
print('Note: this just a demo and does not suggest which sampling is superior')



In the figures above from left to right, they are generated from Un-annealed ODE-like, Annealed ODE-like, Un-annealed Langevin dynamics and Annealed Langevin dynamics.
Note: this just a demo and does not suggest which sampling is superior.
3. Critical components for score-based diffusion in SE(3)
In the previous example, we use a (ground truth) score model compute_score_image to generate (the same) images by following the score. This example implies that if we want to generalize score-matching diffusion model, we need to define the following on SO(3):
- Scale,
scale_rotations - Add, composition of rotations,
R1 @ R2orcompose_rotvec(v1, v2) - Gaussian. IGSO3
Further, it also depends on the representation we are working on. We will working with the vector or axis-angle representation for rotation in SO(3) and translation vector in \(R^3\). Together, we have a 6-degree-of-freedom representation for SE(3) data. T
# Sampling from a SE-3 Gaussian Distribution
def sample_from_se3_gaussian(x_tar, R_tar, std):
'''
x_tar: translational mean, (..., 3)
R_tar: rotational mean, (..., 3, 3)
std: standard deviation, (..., )
'''
x_eps = std[:, None] * torch.randn_like(x_tar)
theta_eps = std[:, None] * torch.randn_like(x_tar)
rot_eps = SO3().exp_map(theta_eps).to_matrix()
_x = x_tar + x_eps # compose the translation
_R = R_tar @ rot_eps # compose the rotation by matrix mutiplication
return _x, _R
# another implementation using theseus built-in randn
def sample_from_se3_gaussian_with_theseus(x_tar, R_tar, std):
'''
x_tar: translational mean, (..., 3)
R_tar: rotational mean, (..., 3, 3)
std: standard deviation, (..., )
'''
batch_size = std.shape[0]
x_eps = std[:, None] * torch.randn_like(x_tar)
rot_eps = scale_rotations(SO3.randn(batch_size).to_matrix(), std)
_x = x_tar + x_eps # compose the translation
_R = R_tar @ rot_eps # compose the rotation
return _x, _R
# using the complicated formula of IGSO3
# A helper function construct H from x and R
# H = [[R, x],
# [0, 1]]
def construct_roto_trans(x, R):
assert len(R.shape) == 3
assert x.shape[0] == R.shape[0]
batch_size = x.shape[0]
H = torch.eye(4)[None, ...].repeat(batch_size, 1, 1)
H[:, :3, :3] = R.clone()
H[:, :3, -1] = x.clone()
return Htorch.manual_seed(42)
B = 400
R_mu = SO3.rand(1).to_matrix().repeat(B, 1, 1) # mean rotation
x_mu = torch.randn(1, 3).repeat(B, 1) # mean translation
# rotation and translation combined in the H tensor (..., 4, 4)
H_mu = construct_roto_trans(x_mu, R_mu)
print('An example of SE(3) group element:')
print(H_mu[0:1])
# Visualizations of Gaussians on SO(3) centering at mean rotation with different std
stds = [1.0, 0.8, 0.5, 0.25, 0.1, 0.05]
rots, rots_with_theseus = [], []
for std in stds:
__, R_samples = sample_from_se3_gaussian(x_mu, R_mu, std * torch.ones(B))
__, R_samples_with_theseus = sample_from_se3_gaussian_with_theseus(x_mu, R_mu, std * torch.ones(B))
rots.append(R_samples)
rots_with_theseus.append(R_samples_with_theseus)
print('sample_from_se3_gaussian')
fig = plot_rotations(rots, labels=stds)
fig.to_html('se3_gaussian.html')
print('sample_from_se3_gaussian_with_theseus')
fig = plot_rotations(rots_with_theseus, labels=stds)
fig.to_html('se3_gaussian_th.html')
# Outputs:
# An example of SE(3) group element:
# tensor([[[-0.7645, 0.1293, -0.6315, -0.8293],
# [ 0.6156, -0.1438, -0.7748, -1.6137],
# [-0.1910, -0.9811, 0.0303, -0.2147],
# [ 0.0000, 0.0000, 0.0000, 1.0000]]])sample_from_se3_gaussian:
sample_from_se3_gaussian_with_theseus
# A spreadout SE(3) Gaussian, std = 0.4
torch.manual_seed(42)
x_samples, R_samples = sample_from_se3_gaussian(x_mu, R_mu, 0.4 * torch.ones(B))
H = construct_roto_trans(x_samples, R_samples)
H = torch.cat([H, H_mu[0:1]], dim=0)
colors = torch.zeros_like(H[:, :3, -1])
colors[-1, 0] = 1
scene = visualize_grasps(Hs=H, colors=colors.numpy(), show=False)
display.display(scene.show())
# A tight SE(3) Gaussian, std = 0.25
torch.manual_seed(42)
x_samples, R_samples = sample_from_se3_gaussian(x_mu, R_mu, 0.25 * torch.ones(B))
H = construct_roto_trans(x_samples, R_samples)
H = torch.cat([H, H_mu[0:1]], dim=0)
colors = torch.zeros_like(H[:, :3, -1])
colors[-1, 0] = 1
scene = visualize_grasps(Hs=H, colors=colors.numpy(), show=False)
display.display(scene.show())
print('Note that each fork is of the same size!')\(\sigma = 0.40\)
\(\sigma = 0.25\)
Note that each fork is of the same size!
# Evaluate log probability of SE(3) poses in SE(3) Gaussian distribution
def se3_log_probability_normal(x, R, x_tar, R_tar, std, eps=1e-6):
'''
x: translation samples
R: rotation samples
x_tar: mean translation of SE-3 gaussian
R_tar: mean rotation of SE-3 gaussian
std: standard deviation of the SE-3 gaussian
'''
assert x.shape == x_tar.shape
assert R.shape == R_tar.shape
# Mean rotation as theseus object
_R_tar = SO3()
_R_tar.update(R_tar)
# rotation samples as theseus object
_R = SO3()
_R.update(R)
R = _R
# Compute distance in R^3 + SO(3)
# Rotation distance
R_tar_inv = _R_tar.inverse()
dR = SO3()
dR_rot = R_tar_inv.to_matrix() @ R.to_matrix()
dR.update(dR_rot)
dv = dR.log_map() # the vector representation for the rotation difference
# translation distance
dx = (x - x_tar)
# 6D distance
dist = torch.cat((dx, dv), dim=-1)
# compute the log probability up to a constant term, which we don't care
return -.5 * dist.pow(2).sum(-1) / (std.pow(2) + eps)
std = 0.25 * torch.ones(B)
log_prob = se3_log_probability_normal(x_samples, R_samples, x_mu, R_mu, std)
# probability of each sample comping from the SE-3 samples
colors = torch.zeros((B + 1, 3))
colors[:-1, 1] = (log_prob - log_prob.min())/(log_prob.max() - log_prob.min())
colors[:-1, 0] = 1. - (log_prob - log_prob.min())/(log_prob.max() - log_prob.min())
scene = visualize_grasps(Hs=H, colors=colors.numpy(), show=False)
display.display(scene.show())
print('These forks are colored by the log probability, the greener the greater the logp < 0')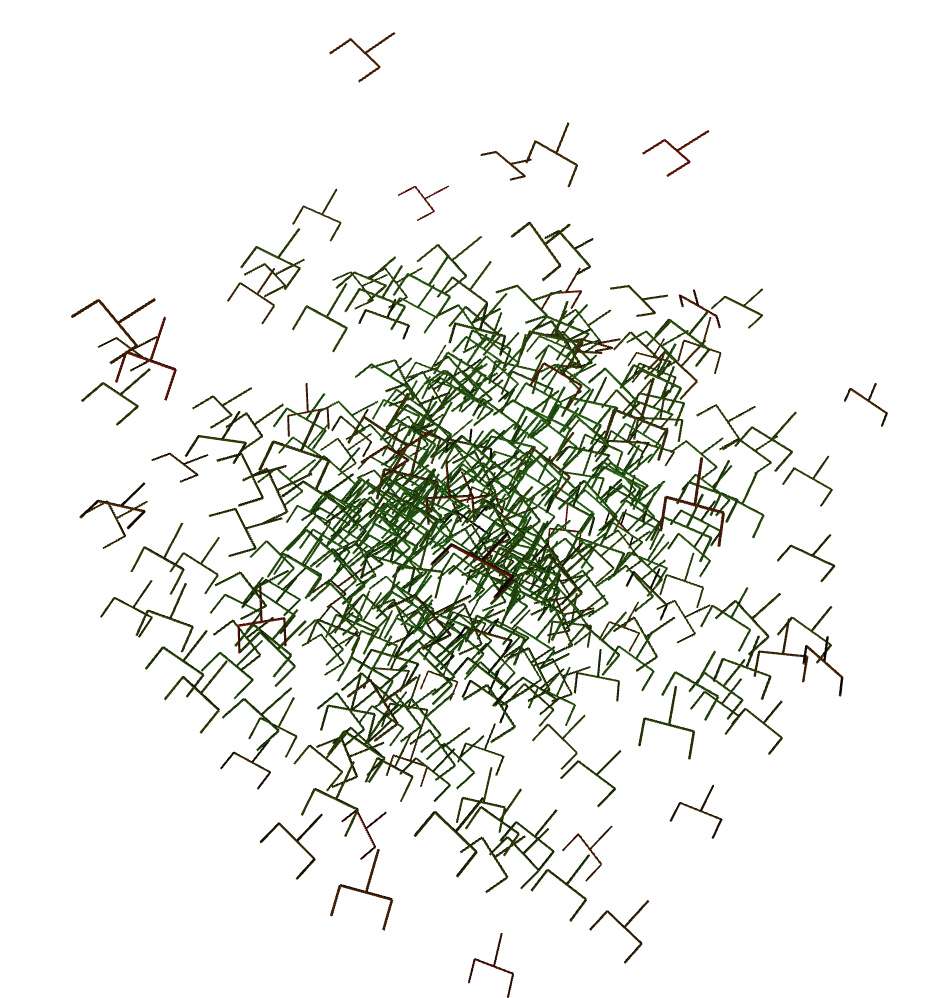
These forks are colored by the log probability, the greener the greater the \(logp < 0\)
We have calculated the SE(3) log probability density evaluated at \(x(t)\) or trans: \(x(t)\) and rot: \(R(t)\), up to a constant term. We now want to do 2 things:
- Compute the score, i.e. take the gradient of log probability wrt \(x(t)\) and \(R(t)\)
This can be done analytically with the IGSO3 formula and the quotient rule as done in FrameDiff (Yim et al ICLR 2023). Or can be done via theseus’s compatibility with autodiff. We will use the latter in this notebook.
The score is of the same dimension of the data, which has 6 degrees of freedom. We will denote as \(v = (x, y, z, w_x, w_y, w_z)\).
- Move one step in the direction of the score.
This involves doing integration of small steps on the SO(3) manifold, converting the small axis-angle vector (in so(3)) to rotation matrix (in SO(3)) step by step as illustrated in the following figure.

# Move an SE(3) pose given the score of a Gaussian Distribution in SE(3)
# compute the SE(3) scores
def se3_score_normal(x, R, x_tar, R_tar, std):
'''
x: translational samples. (..., 3)
R: rotational samples. (..., 3, 3)
v: se3 scores. (..., 6)
'''
# theseus object
_R = SO3()
_R.update(R)
R = _R
# construct trainable 6D vector
v = R.log_map()
x_theta = torch.cat((x, v), dim=-1)
x_theta.requires_grad_(True)
# assign components from 6D vector
# looks redundant but this preserves the gradient hook and computation graph
x = x_theta[..., :3]
R = SO3.exp_map(x_theta[..., 3:])
# compute log probability with gradient hooked tensors
d = se3_log_probability_normal(x, R, x_tar, R_tar, std)
v = grad(d.sum(), x_theta, only_inputs=True)[0]
return v
# (x, R) + v
def step(x, R, v):
# compose rotations
rot = SO3.exp_map(v[..., 3:]).to_matrix()
R_1 = R @ rot
# compose translations
x_1 = x + v[...,:3]
return x_1, R_1
# a helper function for scaling the roto-translation vector
# using the proper scaling of rotations
def scale_roto_trans_vec(v, scale):
assert v.shape[0] == scale.shape[0]
trans, roto = v[..., :3], v[..., 3:]
# roto vector scaling
R = SO3().exp_map(roto).to_matrix()
R = scale_rotations(R, scale)
roto = SO3(tensor=R).log_map()
# trans vector scaling
trans = trans * scale[..., None]
return torch.cat((trans, roto), dim=-1)
# follow se3 scores, keeping intermediate states and outputs the scene
def follow_score_se3(x_truth, R_truth, std_min, std_max, T=2000, dt=0.001, noise_on=False, naive_scale=True, seed=41):
assert x_truth.shape[0] == R_truth.shape[0] == 1
H_truth = construct_roto_trans(x_truth, R_truth)
# start with some random position and rotation in SE(3)
torch.manual_seed(seed)
R = SO3.rand(1).to_matrix() # the prior is USO3
x = torch.randn(1, 3) # the prior is Normal
# std schedules
stds = torch.linspace(std_max, std_min, T + 1)
sqrt2_dt = np.sqrt(2 * dt)
# init SE3 components
H_trj = torch.zeros(0, 4, 4)
# following the scores
for i in tqdm(range(T + 1)):
H0 = construct_roto_trans(x.detach(), R.detach())
H_trj = torch.cat((H_trj, H0), dim=0)
v = se3_score_normal(x, R, x_truth, R_truth, std=torch.tensor([stds[i]]))
_s = v * dt if naive_scale else scale_roto_trans_vec(v, torch.tensor([dt]))
if noise_on and i < T:
z = torch.randn_like(v)
_s += sqrt2_dt * z if naive_scale else scale_roto_trans_vec(z, torch.tensor([sqrt2_dt]))
# one step following the score
x, R = step(x, R, _s)
# for vis
H = torch.cat((H_trj, H_truth), dim=0)
colors = torch.zeros_like(H[:,:3,-1])
colors[:-1, 1] = torch.linspace(0, 1, H_trj.shape[0])
colors[:-1, 2] = 1 - torch.linspace(0, 1, H_trj.shape[0])
colors[-1,0] = 1
scene = visualize_grasps(Hs=H, colors=colors.numpy(), show=False)
return H, sceneprint('Un-annealed ODE-like with large std...')
__, scene = follow_score_se3(x_mu[:1, ...], R_mu[:1, ...], 0.5, 0.5, noise_on=False)
display.display(scene.show())
print('Un-annealed ODE-like with small std...')
__, scene = follow_score_se3(x_mu[:1, ...], R_mu[:1, ...], 0.1, 0.1, noise_on=False)
display.display(scene.show())
print('Annealed ODE-like')
__, scene = follow_score_se3(x_mu[:1, ...], R_mu[:1, ...], 0.1, 5.0, noise_on=False)
display.display(scene.show())
print('Un-annealed Langevin dyanmics with large std...')
__, scene = follow_score_se3(x_mu[:1, ...], R_mu[:1, ...], 0.5, 0.5, noise_on=True)
display.display(scene.show())
print('Un-annealed Langevin dyanmics with small std...')
__, scene = follow_score_se3(x_mu[:1, ...], R_mu[:1, ...], 0.1, 0.1, noise_on=True)
display.display(scene.show())
print('Annealed Langevin dyanmics')
__, scene = follow_score_se3(x_mu[:1, ...], R_mu[:1, ...], 0.1, 5.0, noise_on=True)
display.display(scene.show())4. Training SE(3) Diffusion model
Toy example 1
Here we want to train a model that generates a 2 SE(3) elements.
# Training a Toy SE(3) Diffusion Model
# getting the data
def get_sample_from_data(B=100):
x_data = torch.Tensor([[0.3, 0.3, 0.3],
[-0.5, 1.2, -0.7]])
theta_data = torch.Tensor([[0., 0.0, 0.0],
[-0.3, 1.2, -0.4]])
R_data = SO3().exp_map(theta_data).to_matrix()
idx = torch.randint(0, 2, (B, ))
_x = x_data[idx, ...]
_R = R_data[idx, ...]
return _x, _R
# defines the scheduling of std schedule with SDE with no drift
# this is the variance schedule we chose
def marginal_prob_std(t, sigma=0.5):
return torch.sqrt((sigma ** (2 * t) - 1.) / (2. * np.log(sigma)))
# Define the layer and model
# Time step embedding
class GaussianFourierProjection(nn.Module):
'''Gaussian random features for encoding time steps.
'''
def __init__(self, embed_dim, scale=30.):
super().__init__()
self.W = nn.Parameter(torch.randn(embed_dim // 2) * scale, requires_grad=False) # fixed random projection
def forward(self, x):
x_proj = torch.einsum('...,b->...b', x, self.W) * 2 * torch.pi
return torch.cat([torch.sin(x_proj), torch.cos(x_proj)], dim=-1)
# Naive all linears
class NaiveSE3DiffusionModel(nn.Module):
'''Basic NN with linears
input the noised x (B, 3) and R (B, 3, 3) at time t (B, )
output the predicted scores v (B, 6)
'''
def __init__(self):
super().__init__()
input_size = 12 # we take a translation and flattened rotation
# one can do a 6-dim where we take the Log of the rotation
enc_dim = 128
output_size = 6
self.network = nn.Sequential(
nn.Linear(2 * enc_dim, 128),
nn.ReLU(),
nn.Linear(128, 256),
nn.ReLU(),
nn.Linear(256, output_size)
)
# Time Embedings Encoder
self.time_embed = nn.Sequential(
GaussianFourierProjection(embed_dim=enc_dim),
nn.Linear(enc_dim, enc_dim),
nn.SiLU(),
)
self.x_embed = nn.Sequential(
nn.Linear(input_size, enc_dim),
nn.SiLU(),
)
def forward(self, x, R, t):
std = marginal_prob_std(t)
x_R_input = torch.cat((x, R.reshape(R.shape[0], -1)), dim=-1)
z = self.x_embed(x_R_input)
z_time = self.time_embed(t)
z_in = torch.cat((z, z_time),dim=-1)
v = self.network(z_in)
# the 1/v scaling is necessary for numerical stability
# as we don't want the NN to predict scores say, from 0.001 to 100.0
return v / (std[..., None].pow(2))
# Training
model = NaiveSE3DiffusionModel()
optimizer = torch.optim.AdamW(model.parameters(), lr=0.0005)
K = 1000
B = 500
EPS = 1e-3
loss_trj = torch.zeros(0)
for k in tqdm(range(K)):
t = (1 - EPS) * torch.rand(B) + EPS # t ~ 0 will cause numerical instability
std = marginal_prob_std(t) # compute scheuling of std at t=t, increasing with t
x, R = get_sample_from_data(B) # batch samples
x_eps, R_eps = sample_from_se3_gaussian(x, R, std) # noised samples
v_tar = se3_score_normal(x_eps, R_eps, x_tar=x, R_tar=R, std=std) # estimate scores of noised samples
v_pred = model(x_eps, R_eps, t) # predicted scores from model
loss = std ** 2 * (((v_pred - v_tar) ** 2).sum(-1)) # score matching loss
loss_trj = torch.cat((loss_trj, loss.mean().detach()[None]), dim=0)
# Backward pass and optimization
optimizer.zero_grad()
loss.mean().backward()
optimizer.step()
plt.plot(loss_trj)
plt.show()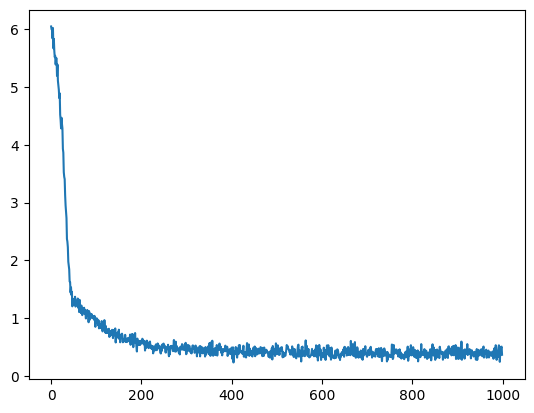
# Given the model run reverse sampling using the step function we defined
def sample_se3(model, data_gen, B=B, T=1000, dt=0.001, eps=1e-3, noise_on=False, naive_scale=True, seed=37):
# random starting point
torch.manual_seed(seed)
R0 = SO3.rand(B).to_matrix()
x0 = torch.randn(B, 3)
for t in range(T):
k = (T - t)/T + eps
v = model(x0, R0, t=k * torch.ones(B))
_s = v * dt if naive_scale else scale_roto_trans_vec(v, dt * torch.ones(B, ))
if noise_on and t != T - 1:
z = torch.randn_like(v)
_s += np.sqrt(2 * dt) * z if naive_scale else scale_roto_trans_vec(z, np.sqrt(2 * dt) * torch.ones(B, ))
x0, R0 = step(x0, R0, _s)
# generated samples (red)
H_gen = construct_roto_trans(x0.detach(), R0.detach())
colors_gen = torch.zeros((B, 3))
colors_gen[:,0] = 1
# real samples (green)
xd, Rd = data_gen(B=10)
H_dat = construct_roto_trans(xd, Rd)
colors_dat = torch.zeros((10, 3))
colors_dat[:,1] = 1
H = torch.cat((H_gen, H_dat), dim=0)
c = torch.cat((colors_gen, colors_dat), dim=0)
scene = visualize_grasps(Hs=H, colors=c.numpy(), show=False)
return H_gen, sceneprint('Sampling from trained model by following score prediction')
__, scene = sample_se3(model, get_sample_from_data, noise_on=False)
display.display(scene.show())
print('Sampling from trained model by Langevin dynamics')
H, scene = sample_se3(model, get_sample_from_data, noise_on=True)
display.display(scene.show())
plot_rotations([R_dummy] + [H[..., :3, :3]] + [get_sample_from_data(10)[1]])2. Toy example 2
Here we want to train a model that generates a 4 SE(3) elements.
We’ll later see that the translation part defines unbalanced sampling and tricks to overcome it.
# try different distributions
def get_sample_from_data_2(B=100):
x_data = torch.Tensor([[0.3, 0.3, 0.3],
[-0.5, 1.2, -0.7],
[3.1, 1.3, -2.4], # this one is significantly more further away
[-0.3, 1.2, 0.4]])
theta_data = torch.Tensor([[0., 0.0, 0.0],
[-0.3, 1.2, -0.4],
[0.3, -1., -1.4],
[-0.5, 1.2, 0.7]])
R_data = SO3().exp_map(theta_data).to_matrix()
idx = torch.randint(0, 4, (B,))
_x = x_data[idx, :]
_R = R_data[idx, ...]
return _x, _R
# the same training, just copied and pasted here
model = NaiveSE3DiffusionModel()
optimizer = torch.optim.AdamW(model.parameters(), lr=0.0005)
K = 1000
B = 500
loss_trj = torch.zeros(0)
for k in tqdm(range(K)):
t = (1 - EPS) * torch.rand(B) + EPS
std = marginal_prob_std(t)
x, R = get_sample_from_data_2(B)
x_eps, R_eps = sample_from_se3_gaussian(x, R, std)
v_tar = se3_score_normal(x_eps, R_eps, x_tar=x, R_tar=R, std=std)
v_pred = model(x_eps, R_eps, t)
loss = std ** 2 *(((v_pred - v_tar) ** 2).sum(-1))
loss_trj = torch.cat((loss_trj, loss.mean().detach()[None]), dim=0)
optimizer.zero_grad()
loss.mean().backward()
optimizer.step()
plt.plot(loss_trj)
plt.show()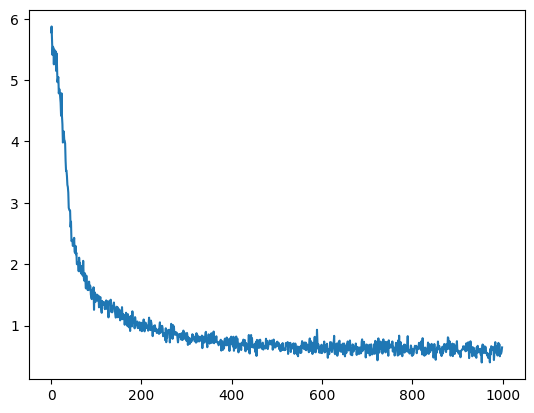
print('Sampling from trained model by following score prediction')
__, scene = sample_se3(model, get_sample_from_data_2, noise_on=False)
display.display(scene.show())
print('Sampling from trained model by Langevin dynamics')
H, scene = sample_se3(model, get_sample_from_data_2, noise_on=True)
display.display(scene.show())
plot_rotations([R_dummy] + [H[..., :3, :3]] + [get_sample_from_data_2(20)[1]])
print()
print('Notice there is a relative sampling bias with respective to the more distant point')
print('This is because for translational sampling, the more distance from the original, the less likely it will be achieved')
print('One can simply make the system of zero COM.')Notice there is a relative sampling bias with respective to the more distant point This is because for translational sampling, the more distance from the original, the less likely it will be achieved One can simply make the system of zero COM.
# Let's redo th training by centering the SE(3) distribution at origin
offset = torch.tensor([0.6500, 1.0000, -0.6000])
model = NaiveSE3DiffusionModel()
optimizer = torch.optim.AdamW(model.parameters(), lr=0.0005)
# again copied and pasted
# should've put this into a function but well..
K = 1000
B = 500
loss_trj = torch.zeros(0)
for k in tqdm(range(K)):
t = (1 - EPS) * torch.rand(B) + EPS
std = marginal_prob_std(t)
x, R = get_sample_from_data_2(B)
# the only difference is the centering
x -= offset
x_eps, R_eps = sample_from_se3_gaussian(x, R, std)
v_tar = se3_score_normal(x_eps, R_eps, x_tar=x, R_tar=R, std=std)
v_pred = model(x_eps, R_eps, t)
loss = std ** 2 *(((v_pred - v_tar) ** 2).sum(-1))
loss_trj = torch.cat((loss_trj, loss.mean().detach()[None]), dim=0)
# Backward pass and optimization
optimizer.zero_grad()
loss.mean().backward()
optimizer.step()
plt.plot(loss_trj)
plt.show()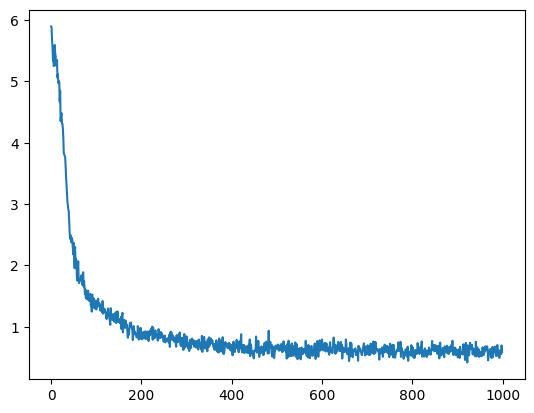
print('Sampling from trained model by Langevin dynamics')
H, scene = sample_se3(model, get_sample_from_data_2, noise_on=True)
display.display(scene.show())
plot_rotations([R_dummy] + [H[..., :3, :3]] + [get_sample_from_data_2(20)[1]])
print()
print('The green forks are from un-centered distribution')
print('Notice that the sampling becomes more balanced by just centering the distribution')
print('There are ways to improve the sampling, like scaling.')
print('Because the roto-translation are coupled, now we have a more balanced samples for the orientation as well.')
print()
print('There are many ways to overcome this, like scaling, sample and recentering, decoupling')The green forks are from un-centered distribution Notice that the sampling becomes more balanced by just centering the distribution There are ways to improve the sampling, like scaling. Because the roto-translation are coupled, now we have a more balanced samples for the orientation as well.
There are many ways to overcome this, like scaling, sample and recentering, decoupling
3. Real example: A protein!
Here, we will use a protein’s backbone frames as ONE single SE(3) distribution and see if our NN and framework can learn it.
We’ll probably define a more complex / deeper model for this task as we know that the underlying distribution is very complicated.
# Let's use a more complicated SE(3) distribution - protein
import pickle
retrain = False # turn this on if you want to train it yourself
# rotation from 3 points using Gram–Schmidt to construct local basis
# a protein local frame can be constructed using C-alpha, N and C
def from_3_points(
p_neg_x_axis: torch.Tensor,
origin: torch.Tensor,
p_xy_plane: torch.Tensor,
eps: float = 1e-8
):
"""
Implements algorithm 21. Constructs transformations from sets of 3
points using the Gram-Schmidt algorithm.
Args:
p_neg_x_axis: [*, 3] coordinates
origin: [*, 3] coordinates used as frame origins
p_xy_plane: [*, 3] coordinates
eps: Small epsilon value
Returns:
A transformation object of shape [*]
"""
p_neg_x_axis = torch.unbind(p_neg_x_axis, dim=-1)
origin = torch.unbind(origin, dim=-1)
p_xy_plane = torch.unbind(p_xy_plane, dim=-1)
e0 = [c1 - c2 for c1, c2 in zip(origin, p_neg_x_axis)]
e1 = [c1 - c2 for c1, c2 in zip(p_xy_plane, origin)]
denom = torch.sqrt(sum((c * c for c in e0)) + eps)
e0 = [c / denom for c in e0]
dot = sum((c1 * c2 for c1, c2 in zip(e0, e1)))
e1 = [c2 - c1 * dot for c1, c2 in zip(e0, e1)]
denom = torch.sqrt(sum((c * c for c in e1)) + eps)
e1 = [c / denom for c in e1]
e2 = [
e0[1] * e1[2] - e0[2] * e1[1],
e0[2] * e1[0] - e0[0] * e1[2],
e0[0] * e1[1] - e0[1] * e1[0],
]
rots = torch.stack([c for tup in zip(e0, e1, e2) for c in tup], dim=-1)
rots = rots.reshape(rots.shape[:-1] + (3, 3))
return rots
# an in-house preprocessed antibody structure
# this should be provided with the notebook
data = pickle.load(open('./AFQ82415.pkl', 'rb'))
coordinates = torch.from_numpy(data['atom_positions'][:101, :, :3]) # N, Ca, C
rots = from_3_points(coordinates[:, 0], coordinates[:, 1], coordinates[:, 2])
trans = coordinates[..., 1, :] / 10. # we need to scale this in order to get a more balanced samples
p = construct_roto_trans(trans, rots)# interpolate between residues to augment the data
t1, t2 = trans[:-1, ...], trans[1:, ...]
r1, r2 = rots[:-1, ...], rots[1:, ...]
for w in torch.linspace(0.1, 1.0, 20):
t = w * t1 + (1. - w) * t2
r = slerp(r2, r1, torch.ones(100) * w)
p = torch.cat((p, construct_roto_trans(t, r)))
scene = visualize_grasps(p, show=False)
display.display(scene.show())# get samples from backbone
prot = construct_roto_trans(trans, rots)
print(prot.shape)
def get_sample_from_protein(prot=prot, B=500):
n = prot.shape[0]
idx = torch.randint(0, n, (B, ))
H = prot[idx, ...]
_x = H[:, :3, -1]
_R = H[:, :3, :3]
return _x, _R
# a deeper model
# We just naively stacked more Linears in between
class NaiveSE3DiffusionModel2(nn.Module):
def __init__(self):
super().__init__()
input_size = 12
enc_dim = 128
output_size = 6
self.network1 = nn.Sequential(
nn.Linear(2*enc_dim, 128),
nn.ReLU(),
nn.Linear(128, 256),
nn.ReLU(),
nn.Linear(256, 256),
nn.ReLU(),
nn.Linear(256, 256),
nn.ReLU(),
nn.Linear(256, 256),
nn.ReLU(),
nn.Linear(256, enc_dim)
)
self.network2 = nn.Sequential(
nn.Linear(2*enc_dim, 128),
nn.ReLU(),
nn.Linear(128, 256),
nn.ReLU(),
nn.Linear(256, 256),
nn.ReLU(),
nn.Linear(256, 256),
nn.ReLU(),
nn.Linear(256, 256),
nn.ReLU(),
nn.Linear(256, output_size)
)
## Time Embedings Encoder ##
self.time_embed = nn.Sequential(
GaussianFourierProjection(embed_dim=enc_dim),
nn.Linear(enc_dim, enc_dim),
nn.SiLU(),
)
self.x_embed = nn.Sequential(
nn.Linear(input_size, enc_dim),
nn.SiLU(),
)
def marginal_prob_std(self, t, sigma=0.5):
return torch.sqrt((sigma ** (2 * t) - 1.) / (2. * np.log(sigma)))
def forward(self, x, R, t):
std = self.marginal_prob_std(t)
x_R_input = torch.cat((x, R.reshape(R.shape[0], -1)), dim=-1)
z = self.x_embed(x_R_input)
z_time = self.time_embed(t)
z_in = torch.cat((z, z_time),dim=-1)
z = self.network1(z_in)
z_in = torch.cat((z, z_time),dim=-1)
v = self.network2(z_in)
return v/(std[:,None].pow(2))if retrain:
torch.manual_seed(42)
model = NaiveSE3DiffusionModel2()
optimizer = torch.optim.AdamW(model.parameters(), lr=0.0005)
K = 30000
B = 1000
loss_trj = torch.zeros(0)
for k in tqdm(range(K)):
t = (1 - EPS) * torch.rand(B) + EPS # t ~ 0 will cause numerical instability
std = marginal_prob_std(t) # compute scheuling of std at t=t, increasing with t
x, R = get_sample_from_protein(B=B) # batch samples
x_eps, R_eps = sample_from_se3_gaussian(x, R, std) # noised samples
v_tar = se3_score_normal(x_eps, R_eps, x_tar=x, R_tar=R, std=std) # estimate scores of noised samples
v_pred = model(x_eps, R_eps, t) # predicted scores from model
loss = std ** 2 * (((v_pred - v_tar) ** 2).sum(-1)) # score matching loss
loss_trj = torch.cat((loss_trj, loss.mean().detach()[None]), dim=0)
# Backward pass and optimization
optimizer.zero_grad()
loss.mean().backward()
optimizer.step()
plt.plot(loss_trj)
plt.show()
# torch.save(model.state_dict(), 'model.pt')if not retrain:
model = NaiveSE3DiffusionModel2()
model.load_state_dict(torch.load('model.pt'))
H, __ = sample_se3(model, get_sample_from_protein, noise_on=False, T=1000, B=500)
H2, __ = sample_se3(model, get_sample_from_protein, noise_on=False, T=1000, B=500)
H = torch.cat((H, H2), dim=0)
colors = torch.zeros((1101, 3))
colors[:1000, 0] = 1.0
colors[1000:, 1] = 1.0
scene = visualize_grasps(torch.cat((H, prot), dim=0), colors=colors.numpy(), show=False)
display.display(scene.show())colors = torch.zeros((H.shape[0] + p.shape[0], 3))
colors[:1000, 0] = 1.0
colors[1000:, 1] = 1.0
scene = visualize_grasps(torch.cat((H, p), dim=0), colors=colors.numpy(),show=False)
display.display(scene.show())
print('Green is the grund truth')5. Final remarks
The SO(3) data can be transformed to the corresponding vector (axis angle) representation via the Log_map. Approximating SO(3) is equivalent to approximating the vector representation, at least in this notebook.
Note here that we were doing a single SE(3) distribution, instead of modeling a set or an ordered set of SE(3) distributions, like proteins, robot joints. Therefore, the model is just a bunch of Linear layers. If that’s the problem we are interested, we are modeling the SE(3)\(^N\) distribution, where the interactions between each SE(3) will be critical, the model needs to be SE(3)-equivariant to respect the relative roto-translational information between SE(3)\(^N\). Such model was used in AlphaFold2 (IPA) or Rosettafold (SE(3)-Transformer).
Hope you find this tutorial interesting and useful.